Classify structured data with feature columns
This tutorial demonstrates how to classify structured data (e.g. tabular data in a CSV). We will use Keras to define the model, and feature columns as a bridge to map from columns in a CSV to features used to train the model. This tutorial contains complete code to:
- Load a CSV file using the tidyverse.
- Build an input pipeline to batch and shuffle the rows using tf.data.
- Map from columns in the CSV to features used to train the model using feature columns.
- Build, train, and evaluate a model using Keras.
The Dataset
We will use a small dataset provided by the Cleveland Clinic Foundation for Heart Disease. There are several hundred rows in the CSV. Each row describes a patient, and each column describes an attribute. We will use this information to predict whether a patient has heart disease, which in this dataset is a binary classification task.
Following is a description of this dataset. Notice there are both numeric and categorical columns.
Column Description Feature Type Data Type Age Age in years Numerical integer Sex (1 = male; 0 = female) Categorical integer CP Chest pain type (0, 1, 2, 3, 4) Categorical integer Trestbpd Resting blood pressure (in mm Hg on admission to the hospital) Numerical integer Chol Serum cholestoral in mg/dl Numerical integer FBS (fasting blood sugar > 120 mg/dl) (1 = true; 0 = false) Categorical integer RestECG Resting electrocardiographic results (0, 1, 2) Categorical integer Thalach Maximum heart rate achieved Numerical integer Exang Exercise induced angina (1 = yes; 0 = no) Categorical integer Oldpeak ST depression induced by exercise relative to rest Numerical integer Slope The slope of the peak exercise ST segment Numerical float CA Number of major vessels (0-3) colored by flourosopy Numerical integer Thal 3 = normal; 6 = fixed defect; 7 = reversable defect Categorical string Target Diagnosis of heart disease (1 = true; 0 = false) Classification integer
Setup
We will use Keras and TensorFlow datasets.
## ── Attaching packages ─────────────────────────────────────────────────────────────── tidyverse 1.2.1 ──## ✔ ggplot2 3.2.0 ✔ purrr 0.3.2
## ✔ tibble 2.1.3 ✔ dplyr 0.8.3
## ✔ tidyr 1.0.0 ✔ stringr 1.4.0
## ✔ readr 1.3.1 ✔ forcats 0.4.0## ── Conflicts ────────────────────────────────────────────────────────────────── tidyverse_conflicts() ──
## ✖ dplyr::filter() masks stats::filter()
## ✖ dplyr::lag() masks stats::lag()library(rsample)Read the data
We will use read_csv in order to read the csv file to R.
heart <- pins::pin("https://storage.googleapis.com/applied-dl/heart.csv", "heart")
df <- read_csv(heart)## Parsed with column specification:
## cols(
## age = col_double(),
## sex = col_double(),
## cp = col_double(),
## trestbps = col_double(),
## chol = col_double(),
## fbs = col_double(),
## restecg = col_double(),
## thalach = col_double(),
## exang = col_double(),
## oldpeak = col_double(),
## slope = col_double(),
## ca = col_double(),
## thal = col_character(),
## target = col_double()
## )## Observations: 303
## Variables: 14
## $ age <dbl> 63, 67, 67, 37, 41, 56, 62, 57, 63, 53, 57, 56, 56, 44,…
## $ sex <dbl> 1, 1, 1, 1, 0, 1, 0, 0, 1, 1, 1, 0, 1, 1, 1, 1, 1, 1, 0…
## $ cp <dbl> 1, 4, 4, 3, 2, 2, 4, 4, 4, 4, 4, 2, 3, 2, 3, 3, 2, 4, 3…
## $ trestbps <dbl> 145, 160, 120, 130, 130, 120, 140, 120, 130, 140, 140, …
## $ chol <dbl> 233, 286, 229, 250, 204, 236, 268, 354, 254, 203, 192, …
## $ fbs <dbl> 1, 0, 0, 0, 0, 0, 0, 0, 0, 1, 0, 0, 1, 0, 1, 0, 0, 0, 0…
## $ restecg <dbl> 2, 2, 2, 0, 2, 0, 2, 0, 2, 2, 0, 2, 2, 0, 0, 0, 0, 0, 0…
## $ thalach <dbl> 150, 108, 129, 187, 172, 178, 160, 163, 147, 155, 148, …
## $ exang <dbl> 0, 1, 1, 0, 0, 0, 0, 1, 0, 1, 0, 0, 1, 0, 0, 0, 0, 0, 0…
## $ oldpeak <dbl> 2.3, 1.5, 2.6, 3.5, 1.4, 0.8, 3.6, 0.6, 1.4, 3.1, 0.4, …
## $ slope <dbl> 3, 2, 2, 3, 1, 1, 3, 1, 2, 3, 2, 2, 2, 1, 1, 1, 3, 1, 1…
## $ ca <dbl> 0, 3, 2, 0, 0, 0, 2, 0, 1, 0, 0, 0, 1, 0, 0, 0, 0, 0, 0…
## $ thal <chr> "fixed", "normal", "reversible", "normal", "normal", "n…
## $ target <dbl> 0, 1, 0, 0, 0, 0, 1, 0, 1, 0, 0, 0, 1, 0, 0, 0, 0, 0, 0…Split the dataframe into train, validation, and test
We are going to use the rsample package to split the data into train, validation
and test sets.
# first we split between training and testing sets
split <- initial_split(df, prop = 4/5)
train <- training(split)
test <- testing(split)
# the we split the training set into validation and training
split <- initial_split(train, prop = 4/5)
train <- training(split)
val <- testing(split)nrow(train)## [1] 195nrow(val)## [1] 48nrow(test)## [1] 60Create an input pipeline using tfdatasets
Next, we will wrap the dataframes with tfdatasets. This will enable us to use feature columns as a bridge to map from the columns in the dataframe to features used to train the model. If we were working with a very large CSV file (so large that it does not fit into memory), we would use tfdatasets to read it from disk directly. That is not covered in this tutorial.
df_to_dataset <- function(df, shuffle = TRUE, batch_size = 32) {
ds <- df %>%
tensor_slices_dataset()
if (shuffle)
ds <- ds %>% dataset_shuffle(buffer_size = nrow(df))
ds %>%
dataset_batch(batch_size = batch_size)
}Understand the input pipeline
Now that we have created the input pipeline, let’s call it to see the format of the data it returns. We have used a small batch size to keep the output readable.
train_ds %>%
reticulate::as_iterator() %>%
reticulate::iter_next() %>%
str()## List of 14
## $ age :tf.Tensor([54. 60. 52. 66. 54.], shape=(5,), dtype=float32)
## $ sex :tf.Tensor([0. 0. 1. 1. 1.], shape=(5,), dtype=float32)
## $ cp :tf.Tensor([2. 3. 4. 4. 4.], shape=(5,), dtype=float32)
## $ trestbps:tf.Tensor([132. 102. 128. 112. 110.], shape=(5,), dtype=float32)
## $ chol :tf.Tensor([288. 318. 255. 212. 239.], shape=(5,), dtype=float32)
## $ fbs :tf.Tensor([1. 0. 0. 0. 0.], shape=(5,), dtype=float32)
## $ restecg :tf.Tensor([2. 0. 0. 2. 0.], shape=(5,), dtype=float32)
## $ thalach :tf.Tensor([159. 160. 161. 132. 126.], shape=(5,), dtype=float32)
## $ exang :tf.Tensor([1. 0. 1. 1. 1.], shape=(5,), dtype=float32)
## $ oldpeak :tf.Tensor([0. 0. 0. 0.1 2.8], shape=(5,), dtype=float32)
## $ slope :tf.Tensor([1. 1. 1. 1. 2.], shape=(5,), dtype=float32)
## $ ca :tf.Tensor([1. 1. 1. 1. 1.], shape=(5,), dtype=float32)
## $ thal :tf.Tensor([b'normal' b'normal' b'reversible' b'normal' b'reversible'], shape=(5,), dtype=string)
## $ target :tf.Tensor([0. 0. 0. 1. 1.], shape=(5,), dtype=float32)We can see that the dataset returns a list of column names (from the dataframe) that map to column values from rows in the dataframe.
Create the feature spec
We want to train a model to predict the target variable using Keras but, before
that we need to prepare the data. We need to transform the categorical variables
into some form of dense variable, we usually want to normalize all numeric columns too.
The feature spec interface works with data.frames or TensorFlow datasets objects.
Let’s start creating our feature specification:
spec <- feature_spec(train_ds, target ~ .)The first thing we need to do after creating the feature_spec is decide on the variables’ types.
We can do this by adding steps to the spec object.
spec <- spec %>%
step_numeric_column(
all_numeric(), -cp, -restecg, -exang, -sex, -fbs,
normalizer_fn = scaler_standard()
) %>%
step_categorical_column_with_vocabulary_list(thal)The following steps can be used to define the variable type:
-
step_numeric_columnto define numeric variables -
step_categorical_with_vocabulary_listfor categorical variables with a fixed vocabulary -
step_categorical_column_with_hash_bucketfor categorical variables using the hash trick -
step_categorical_column_with_identityto store categorical variables as integers -
step_categorical_column_with_vocabulary_filewhen you have the possible vocabulary in a file
When using step_categorical_column_with_vocabulary_list you can also provide a vocabulary argument
with the fixed vocabulary. The recipe will find all the unique values in the dataset and use it
as the vocabulary.
You can also specify a normalizer_fn to the step_numeric_column. In this case the variable will be
transformed by the feature column. Note that the transformation will occur in the TensorFlow Graph,
so it must use only TensorFlow ops. Like in the example we offer pre-made normalizers - and they will
compute the normalizing function during the recipe preparation.
You can also use selectors like:
-
starts_with(),ends_with(),matches()etc. (from tidyselect) -
all_numeric()to select all numeric variables -
all_nominal()to select all strings -
has_type("float32")to select based on TensorFlow variable type.
Now we can print the recipe:
## ── Feature Spec ────────────────────────────────────────────────────────────────────────────────────────
## A feature_spec with 8 steps.
## Fitted: FALSE
## ── Steps ───────────────────────────────────────────────────────────────────────────────────────────────
## StepCategoricalColumnWithVocabularyList: thal
## StepNumericColumn: age, trestbps, chol, thalach, oldpeak, slope, ca
## ── Dense features ──────────────────────────────────────────────────────────────────────────────────────
## Feature spec must be fitted before we can detect the dense features.After specifying the types of the columns you can add transformation steps. For example you may want to bucketize a numeric column:
spec <- spec %>%
step_bucketized_column(age, boundaries = c(18, 25, 30, 35, 40, 45, 50, 55, 60, 65))You can also specify the kind of numeric representation that you want to use for your categorical variables.
spec <- spec %>%
step_indicator_column(thal) %>%
step_embedding_column(thal, dimension = 2)Another common transformation is to add interactions between variables using crossed columns.
spec <- spec %>%
step_crossed_column(thal_and_age = c(thal, bucketized_age), hash_bucket_size = 1000) %>%
step_indicator_column(thal_and_age)Note that the crossed_column is a categorical column, so we need to also specify what
kind of numeric tranformation we want to use. Also note that we can name the transformed
variables - each step uses a default naming for columns, eg. bucketized_age is the
default name when you use step_bucketized_column with column called age.
With the above code we have created our recipe. Note we can also define the recipe by chaining a sequence of methods:
spec <- feature_spec(train_ds, target ~ .) %>%
step_numeric_column(
all_numeric(), -cp, -restecg, -exang, -sex, -fbs,
normalizer_fn = scaler_standard()
) %>%
step_categorical_column_with_vocabulary_list(thal) %>%
step_bucketized_column(age, boundaries = c(18, 25, 30, 35, 40, 45, 50, 55, 60, 65)) %>%
step_indicator_column(thal) %>%
step_embedding_column(thal, dimension = 2) %>%
step_crossed_column(c(thal, bucketized_age), hash_bucket_size = 10) %>%
step_indicator_column(crossed_thal_bucketized_age)After defining the recipe we need to fit it. It’s when fitting that we compute the vocabulary
list for categorical variables or find the mean and standard deviation for the normalizing functions.
Fitting involves evaluating the full dataset, so if you have provided the vocabulary list and
your columns are already normalized you can skip the fitting step (TODO).
In our case, we will fit the feature spec, since we didn’t specify the vocabulary list for the categorical variables.
spec_prep <- fit(spec)After preparing we can see the list of dense features that were defined:
str(spec_prep$dense_features())## List of 11
## $ age :NumericColumn(key='age', shape=(1,), default_value=None, dtype=tf.float32, normalizer_fn=<function make_python_function.<locals>.python_function at 0x136510ea0>)
## $ trestbps :NumericColumn(key='trestbps', shape=(1,), default_value=None, dtype=tf.float32, normalizer_fn=<function make_python_function.<locals>.python_function at 0x1311f2ea0>)
## $ chol :NumericColumn(key='chol', shape=(1,), default_value=None, dtype=tf.float32, normalizer_fn=<function make_python_function.<locals>.python_function at 0x1311f2158>)
## $ thalach :NumericColumn(key='thalach', shape=(1,), default_value=None, dtype=tf.float32, normalizer_fn=<function make_python_function.<locals>.python_function at 0x1311f2f28>)
## $ oldpeak :NumericColumn(key='oldpeak', shape=(1,), default_value=None, dtype=tf.float32, normalizer_fn=<function make_python_function.<locals>.python_function at 0x1311ec048>)
## $ slope :NumericColumn(key='slope', shape=(1,), default_value=None, dtype=tf.float32, normalizer_fn=<function make_python_function.<locals>.python_function at 0x1311ec0d0>)
## $ ca :NumericColumn(key='ca', shape=(1,), default_value=None, dtype=tf.float32, normalizer_fn=<function make_python_function.<locals>.python_function at 0x1311ec158>)
## $ bucketized_age :BucketizedColumn(source_column=NumericColumn(key='age', shape=(1,), default_value=None, dtype=tf.float32, normalizer_fn=<function make_python_function.<locals>.python_function at 0x136510ea0>), boundaries=(18.0, 25.0, 30.0, 35.0, 40.0, 45.0, 50.0, 55.0, 60.0, 65.0))
## $ indicator_thal :IndicatorColumn(categorical_column=VocabularyListCategoricalColumn(key='thal', vocabulary_list=('fixed', 'normal', 'reversible'), dtype=tf.string, default_value=-1, num_oov_buckets=0))
## $ embedding_thal :EmbeddingColumn(categorical_column=VocabularyListCategoricalColumn(key='thal', vocabulary_list=('fixed', 'normal', 'reversible'), dtype=tf.string, default_value=-1, num_oov_buckets=0), dimension=2, combiner='mean', initializer=<tensorflow.python.ops.init_ops.TruncatedNormal>, ckpt_to_load_from=None, tensor_name_in_ckpt=None, max_norm=None, trainable=True)
## $ indicator_crossed_thal_bucketized_age:IndicatorColumn(categorical_column=CrossedColumn(keys=(VocabularyListCategoricalColumn(key='thal', vocabulary_list=('fixed', 'normal', 'reversible'), dtype=tf.string, default_value=-1, num_oov_buckets=0), BucketizedColumn(source_column=NumericColumn(key='age', shape=(1,), default_value=None, dtype=tf.float32, normalizer_fn=<function make_python_function.<locals>.python_function at 0x136510ea0>), boundaries=(18.0, 25.0, 30.0, 35.0, 40.0, 45.0, 50.0, 55.0, 60.0, 65.0))), hash_bucket_size=10.0, hash_key=None))Build the model
Now we are ready to define our model in Keras. We will use a specialized layer_dense_features that
knows what to do with the feature columns specification.
We also use a new layer_input_from_dataset that is useful to create a Keras input object copying the structure from a data.frame or TensorFlow dataset.
model <- keras_model_sequential() %>%
layer_dense_features(dense_features(spec_prep)) %>%
layer_dense(units = 32, activation = "relu") %>%
layer_dense(units = 1, activation = "sigmoid")
model %>% compile(
loss = loss_binary_crossentropy,
optimizer = "adam",
metrics = "binary_accuracy"
)Train the model
We can finally train the model on the dataset:
history <- model %>%
fit(
dataset_use_spec(train_ds, spec = spec_prep),
epochs = 15,
validation_data = dataset_use_spec(val_ds, spec_prep),
verbose = 2
)## Epoch 1/15
## 39/39 - 1s - loss: 0.6738 - binary_accuracy: 0.6513 - val_loss: 0.0000e+00 - val_binary_accuracy: 0.0000e+00
## Epoch 2/15
## 39/39 - 0s - loss: 0.5535 - binary_accuracy: 0.7692 - val_loss: 0.5300 - val_binary_accuracy: 0.7708
## Epoch 3/15
## 39/39 - 0s - loss: 0.4731 - binary_accuracy: 0.8103 - val_loss: 0.4800 - val_binary_accuracy: 0.7917
## Epoch 4/15
## 39/39 - 0s - loss: 0.4254 - binary_accuracy: 0.8359 - val_loss: 0.4566 - val_binary_accuracy: 0.7917
## Epoch 5/15
## 39/39 - 0s - loss: 0.3922 - binary_accuracy: 0.8410 - val_loss: 0.4395 - val_binary_accuracy: 0.8125
## Epoch 6/15
## 39/39 - 0s - loss: 0.3678 - binary_accuracy: 0.8564 - val_loss: 0.4326 - val_binary_accuracy: 0.7917
## Epoch 7/15
## 39/39 - 0s - loss: 0.3600 - binary_accuracy: 0.8615 - val_loss: 0.4208 - val_binary_accuracy: 0.8125
## Epoch 8/15
## 39/39 - 0s - loss: 0.3469 - binary_accuracy: 0.8615 - val_loss: 0.4167 - val_binary_accuracy: 0.7917
## Epoch 9/15
## 39/39 - 0s - loss: 0.3404 - binary_accuracy: 0.8615 - val_loss: 0.4076 - val_binary_accuracy: 0.7917
## Epoch 10/15
## 39/39 - 0s - loss: 0.3308 - binary_accuracy: 0.8667 - val_loss: 0.4115 - val_binary_accuracy: 0.7917
## Epoch 11/15
## 39/39 - 0s - loss: 0.3109 - binary_accuracy: 0.8615 - val_loss: 0.4136 - val_binary_accuracy: 0.7917
## Epoch 12/15
## 39/39 - 0s - loss: 0.3239 - binary_accuracy: 0.8667 - val_loss: 0.4033 - val_binary_accuracy: 0.7917
## Epoch 13/15
## 39/39 - 0s - loss: 0.3176 - binary_accuracy: 0.8564 - val_loss: 0.4046 - val_binary_accuracy: 0.7917
## Epoch 14/15
## 39/39 - 0s - loss: 0.3171 - binary_accuracy: 0.8615 - val_loss: 0.3979 - val_binary_accuracy: 0.7917
## Epoch 15/15
## 39/39 - 0s - loss: 0.3040 - binary_accuracy: 0.8615 - val_loss: 0.3967 - val_binary_accuracy: 0.7917plot(history)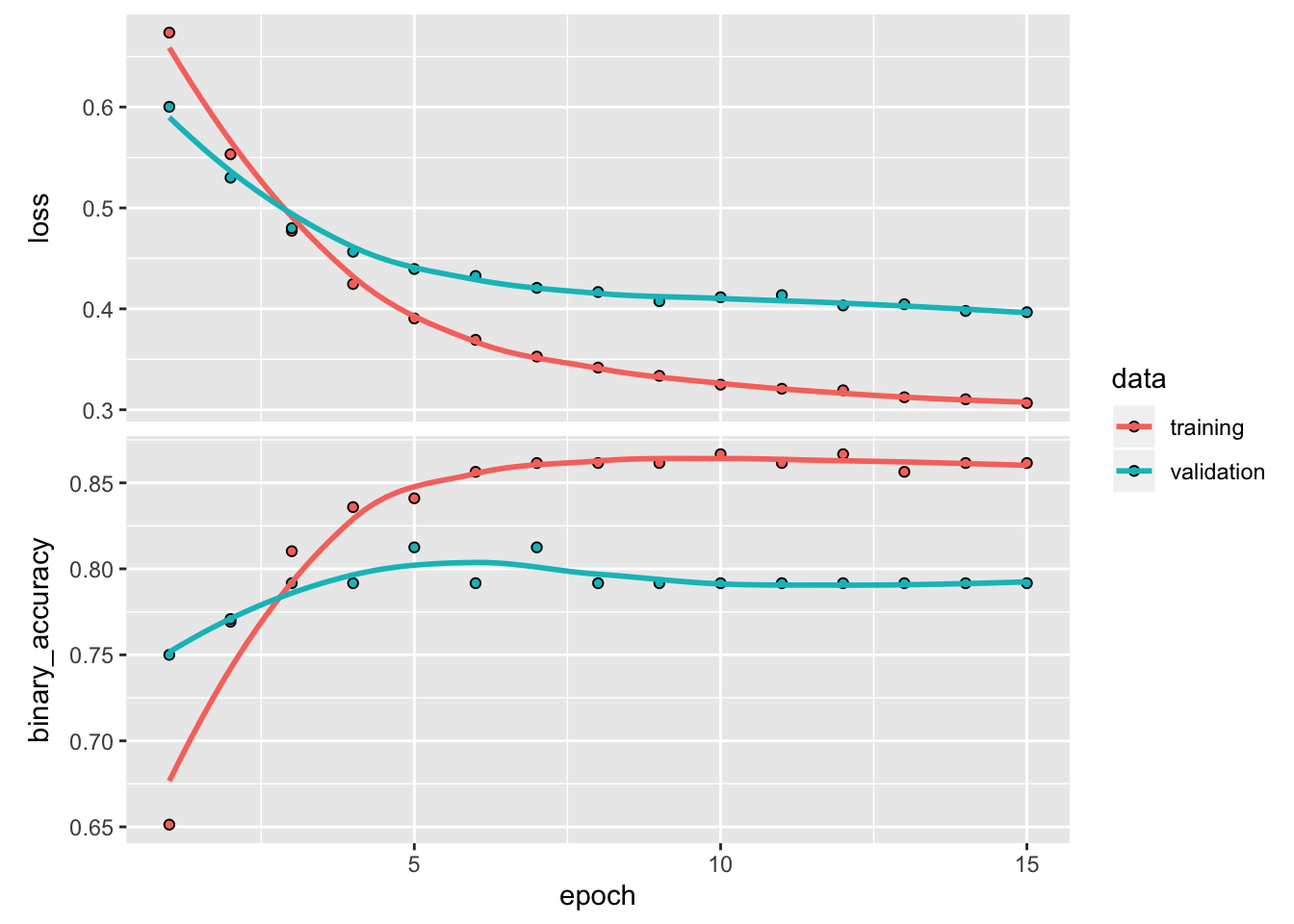
Finally we can make predictions in the test set and calculate performance metrics like the AUC of the ROC curve:
## [1] 0.890538